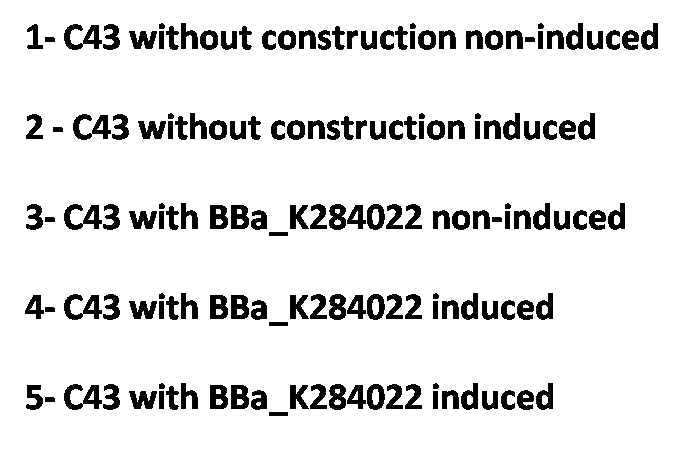Part:BBa_K112806:Experience
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K112806
Lysozymes are widespread in nature. In plants and animals, they constitute a natural defense mechanism against bacterial pathogens. In bacteriophages, lysozymes (called endolysins) locally hydrolyse the peptidoglycan to facilitate penetration of the phage injection apparatus, or induce cell lysis at the end of the phage replication cycle. Bacteria on the other hand also produce lysozymes (called autolysins), that are involved in de novo cell wall synthesis during cell growth and division. The T4 endolysin is 1,4-b-N-acetylmuramidase, cleaving the glycosidic bond between the C-1 of N-acetylmuramic acid (NAM) and the C-4 of N-acetylglucosamine (NAG) in the bacterial peptidoglycan (PG).Therefore, this enzyme can be used whenever the lysis of an bacteria is desired.
By UNICAMP-BRAZIL 2009
User Reviews
UNIQ3fd98952ce2319a2-partinfo-00000000-QINU UNIQ3fd98952ce2319a2-partinfo-00000001-QINU
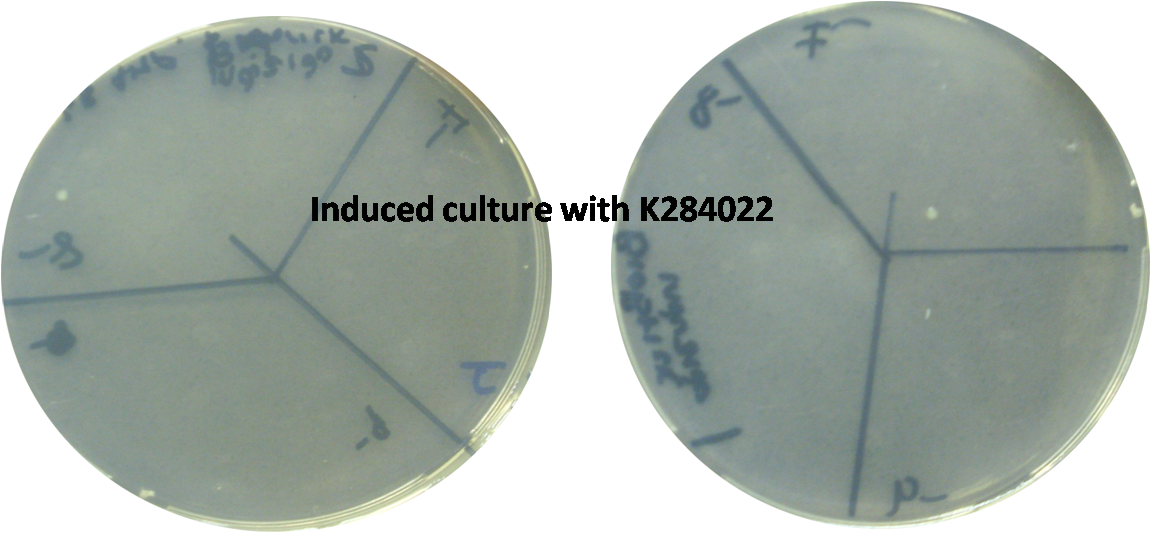
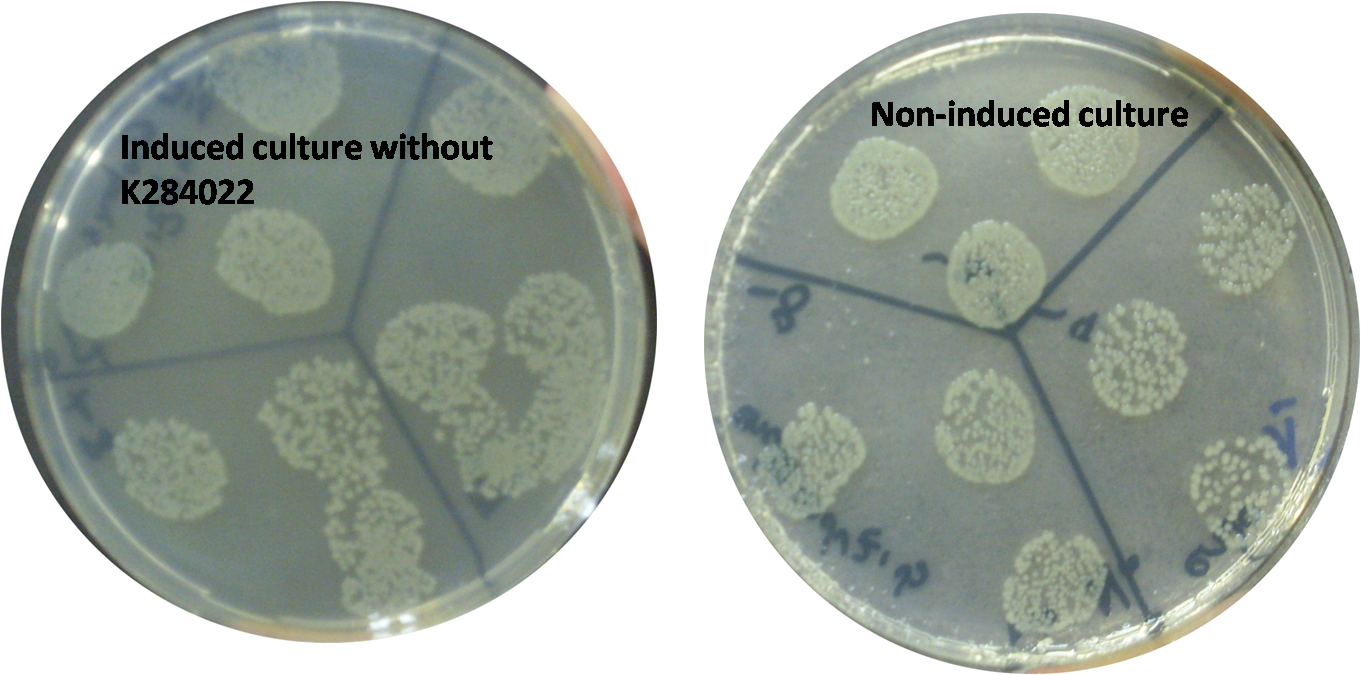
In order to test the T4 endolysin we constructed a new device (BBa_K284022) containing the terminator BBa_B0014 and the part BBa_I746911, which consists of the RBS BBa_B0034 and the T7 promoter. Therefore, the endolysin is controlled by the T7 promoter. Our new device was transformed in E. coli C43 strain, an overexpression strain in which the T7 promoter can be induced by IPTG. A transformed colony was incubated in LB medium with ampicillin at 37ºC overnight. The culture was diluted in new LB medium with ampicillin to an OD=0,2 and incubated at 37ºC until reach an OD=0,8. At this point, the cells were induced with IPTG 1 mM and incubated again for 4 hours. After the end of the induction period the cells were plated in LB agar with ampicillin. The OD measurements showed a decrease on the growth of induced strains, in comparsion with non-induced ones.
Besides that, the death of cells expressing the endolysin was confirmed by the absence of growing colonies on the plates.
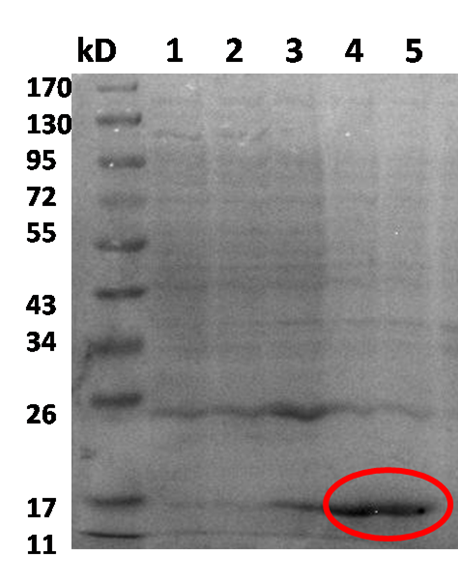
It was also possible to observe the overexpression of T4 endolysin through an SDS-PAGE.
By UNICAMP-BRAZIL 2009
Theoretical Modelling
By GlasgowUni 2011
During our project DISColi, regarding Bio-photolithography in biofilms, we created mathematical models involving T4 endolysin and its rate of diffusion through a biofilm. We are presenting our findings in Figure 1 and Table 1. Also, due to the mode of action of T4 Endolysin the attainable spatial resolution seems to be less than 0.01 mm. For more information, please visit GlasgowUni 2011 Wiki
| (a) |
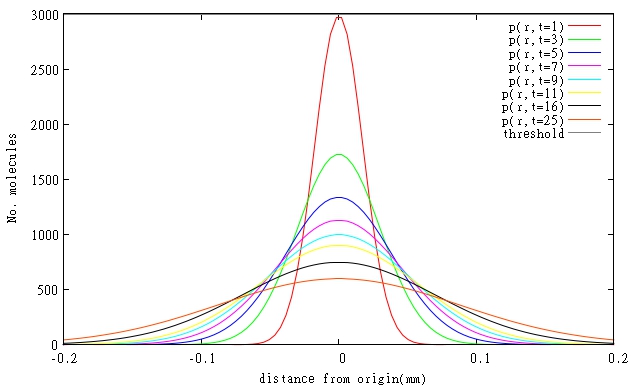 |
| (b) |
 |
| Name of molecule | Length (a.a.) | Molecular weight (g/mol) | Diffusion rate (mm2/min) | Critical concentration (molecules) | Translation rate (molecule/min) | Achieving Critical Concentration (min) |
| T4 endolysin | 164 | 18500 | 1.52*10-4 | 3000 | 57.2 | 52 |